|
Hey whats up, I'm Will. I'm undergrad at UC Berkeley, where I study math and biology. Currently, I work in Adam Arkin's lab on the NASA CUBES project, and I focus on the design and control of microbial communities for agriculture. Before I worked in the Arkin group, I worked in Jay Keasling's lab at the Joint Bio Energy Institute on improving Pseudomonas putida as a microbial chassis for the production of biofuels and commodity chemicals from biomass. Generally, I am interested in the nonlinear dynamics and control of stochastic biological networks. I am fascinated by the application of "structural" controllability to networks like microbiomes and how to control groups of cells when considering them like swarms or distributed systems. Email / Resume / NSF Proposal / Google Scholar / Linkedin |

|
|
|
|
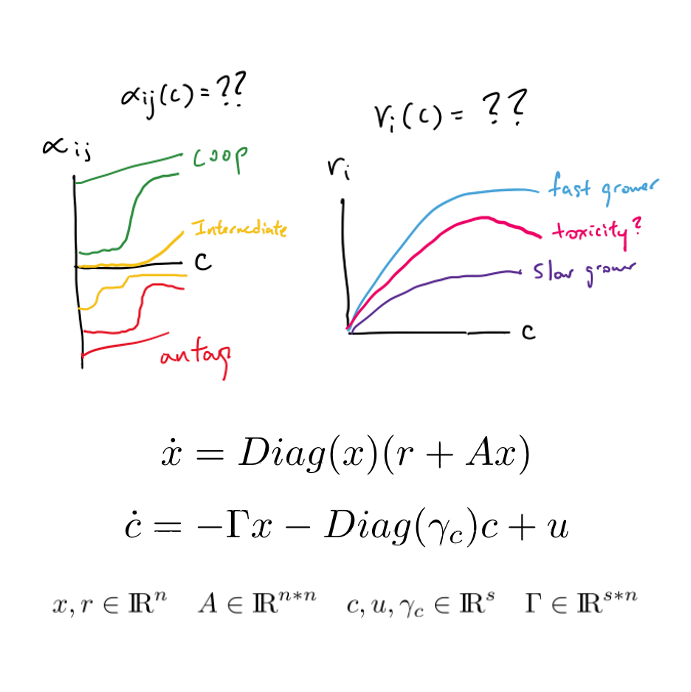
Parameter-varying, Lotka Volterra modeling Senior thesis, in-progress Original Proposal / Grant Presentation / Ppt pdf In this work, I have set out to interrogate the dependence of microbial interactions and growth rates (gLV parameters) on physical and molecular environmental gradients, and thereby, the dependence of community dynamics on the gradients as well. I aim to validate the research by designing an MPC that dictates what temperature and glucose concentration into which a community should be passaged to track a desired diversity. I originally proposed this work as a senior thesis to Adam, and it has since been funded by the UC Berkeley - CNR - SPUR independent research grant. |
|
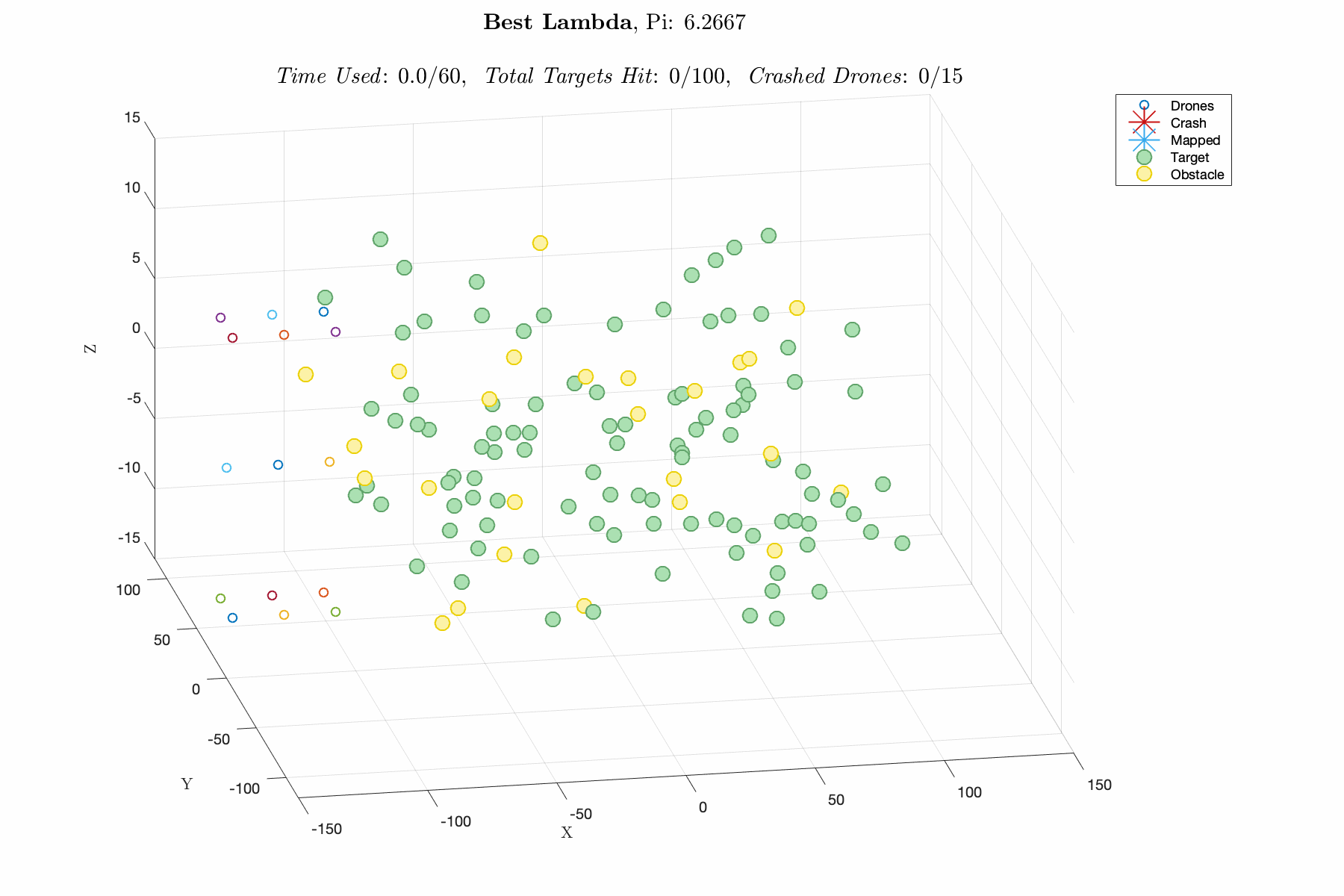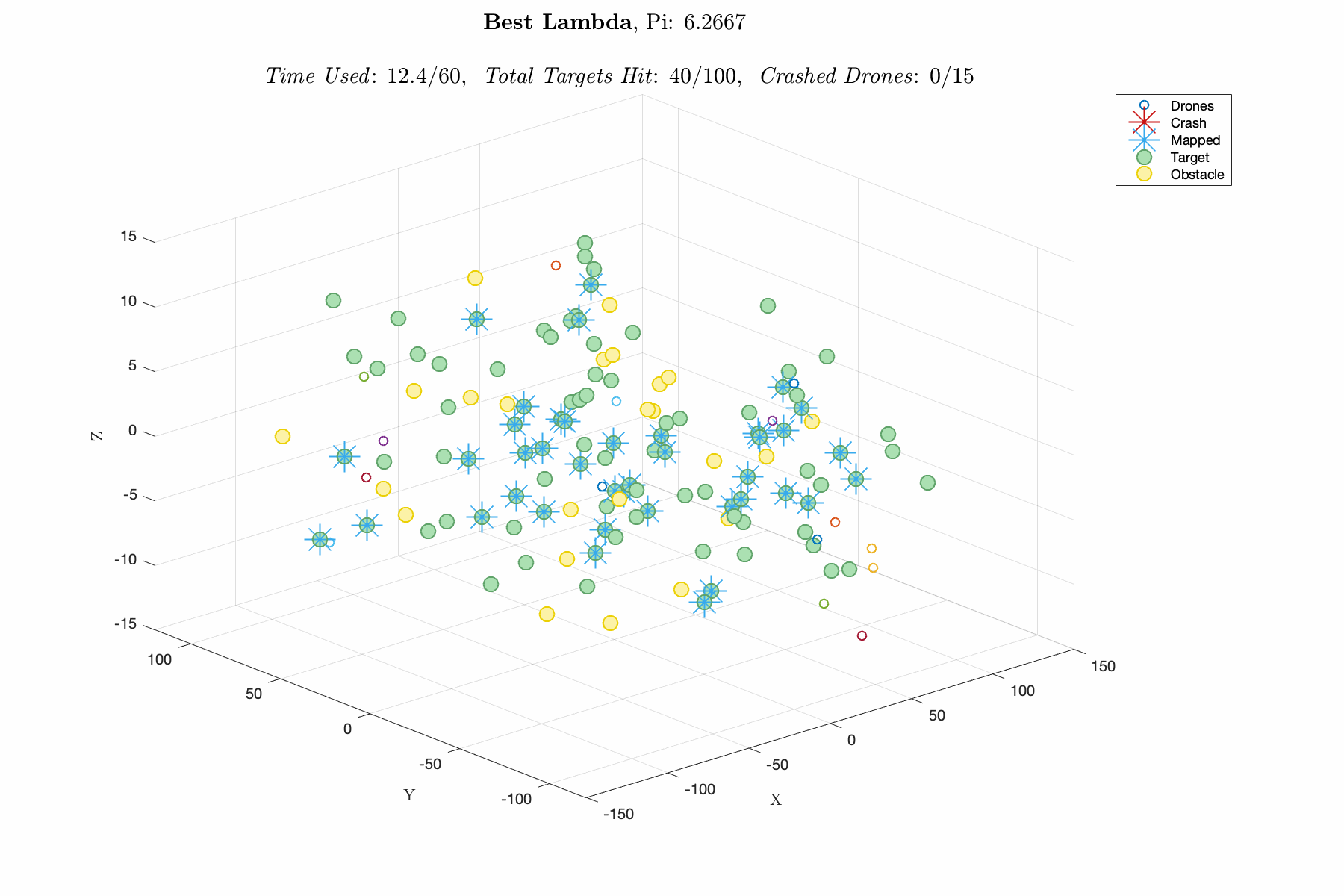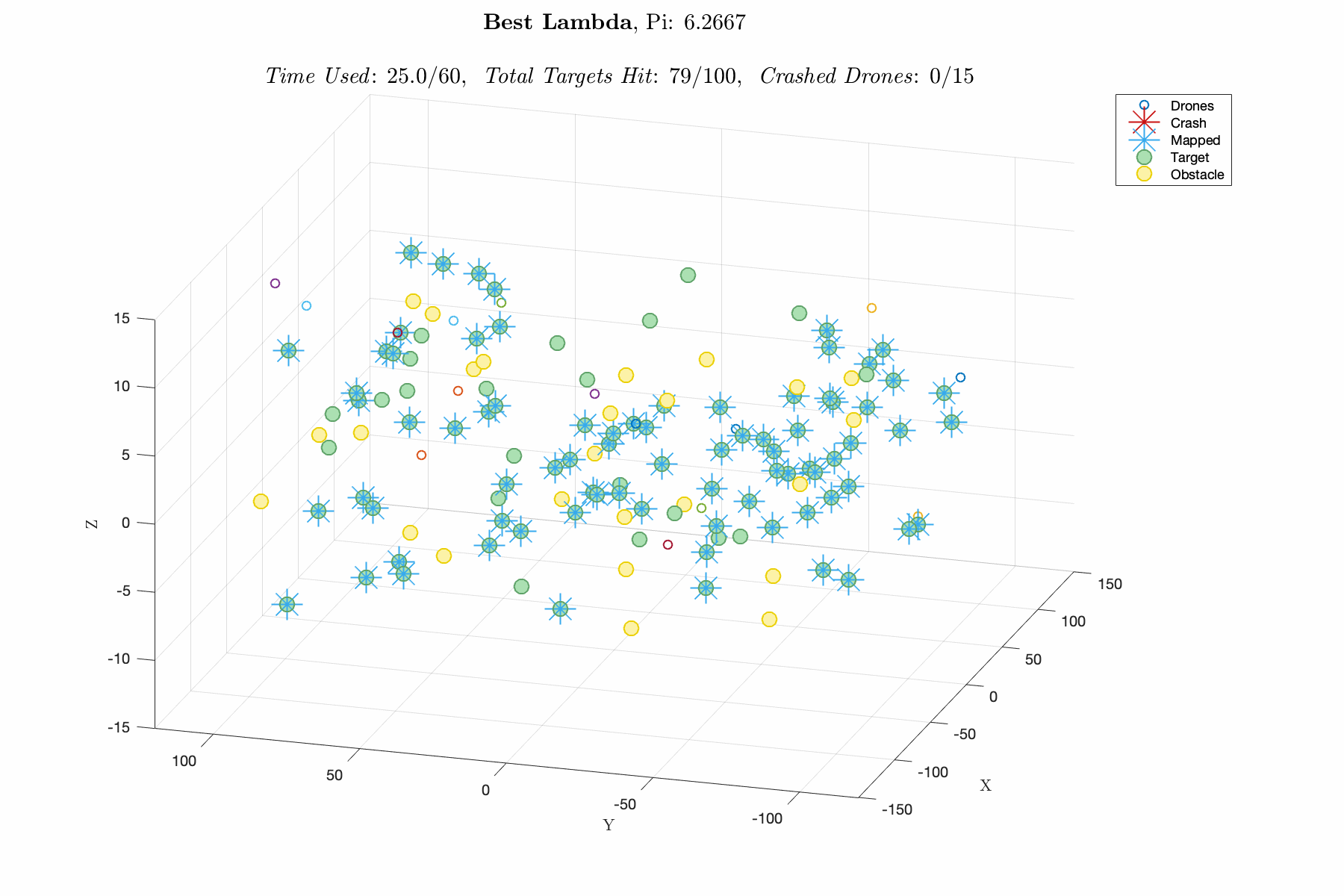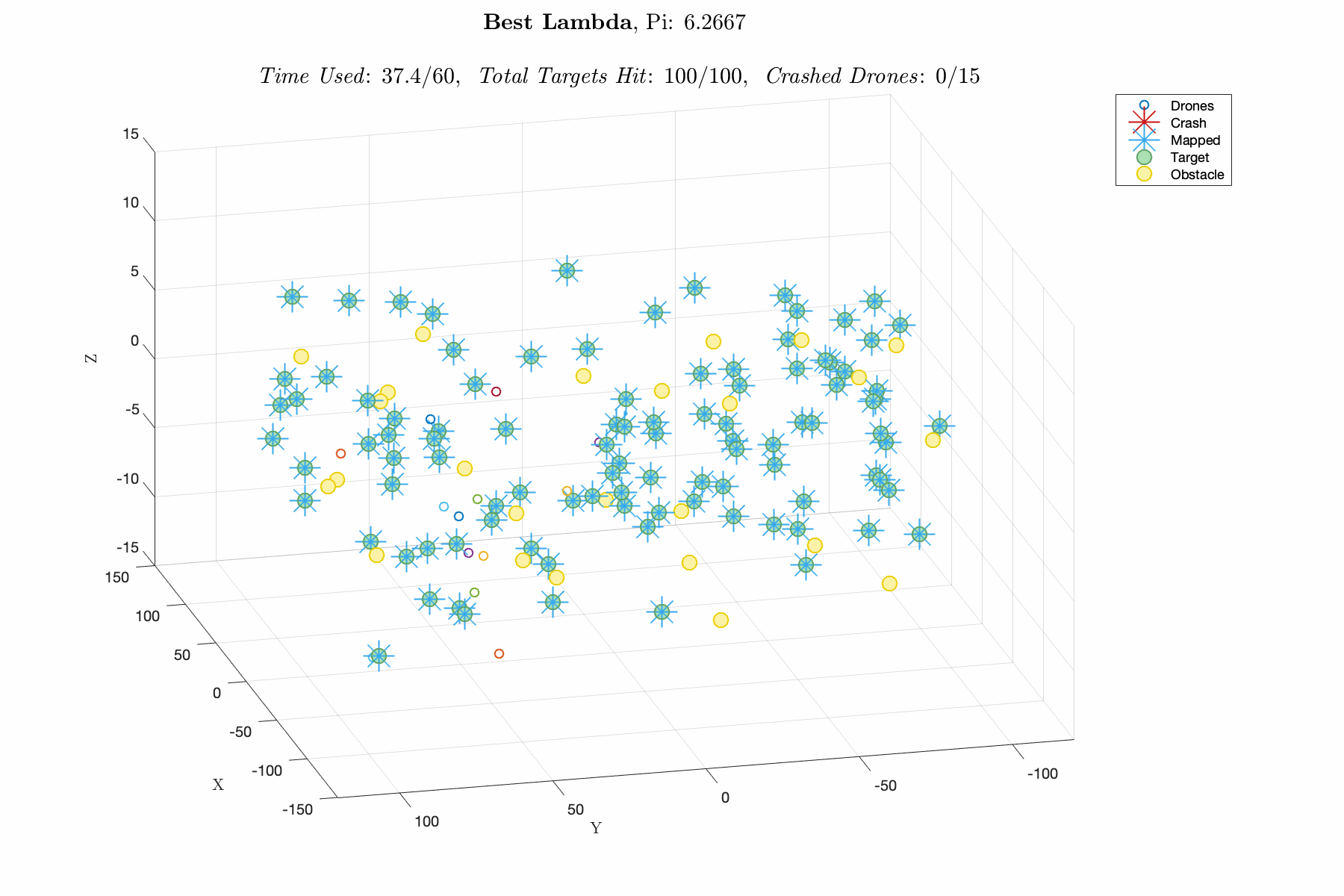
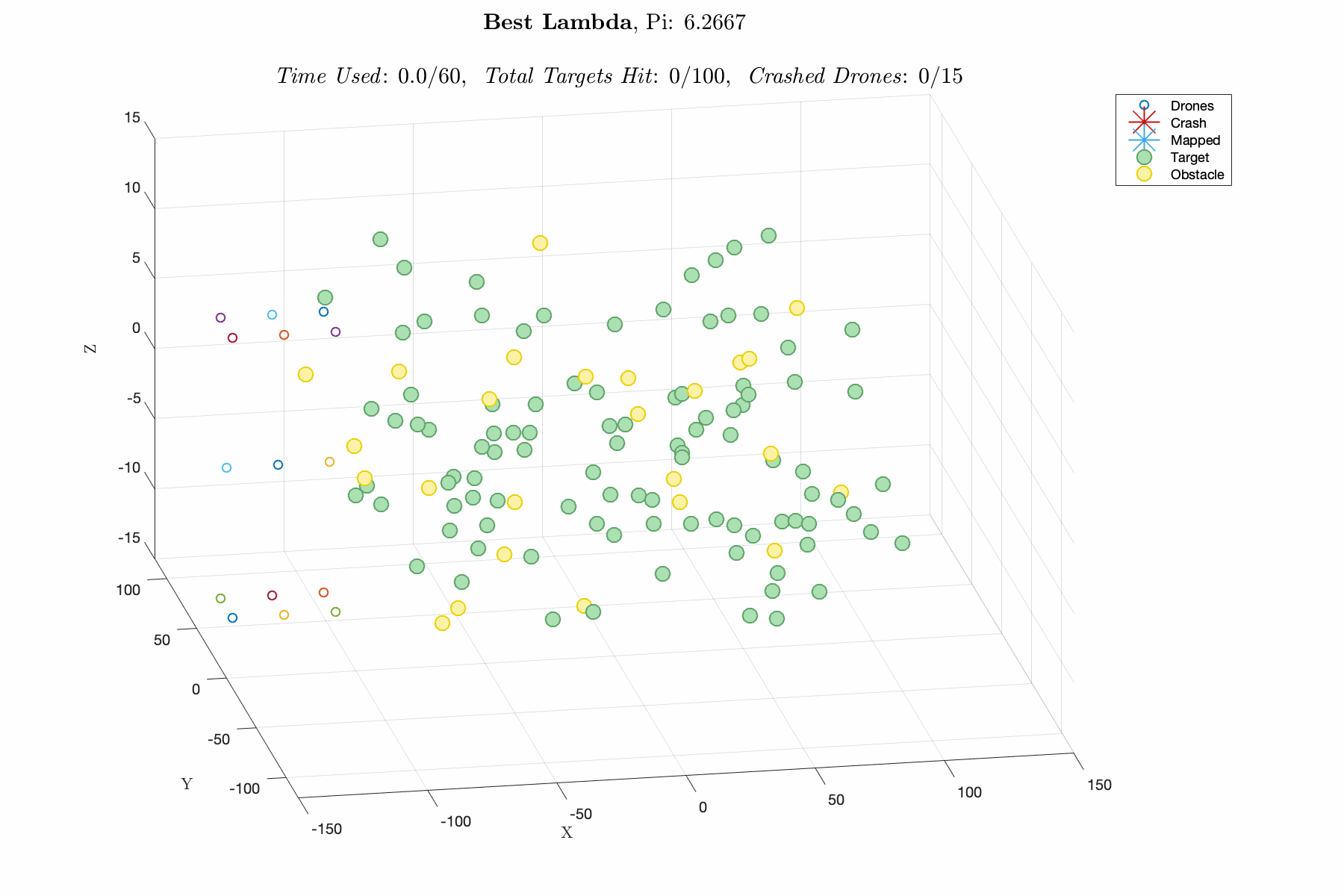
Improving control laws in-silico with a naive ML method E150 Modeling Course Project, Fall 2019 Report / Matlab Script In this project, I utilized a Genetic Algorithm, which breeds top scoring vectors of design parameters, to improve the performance of an autonomous drone swarm tasked with mapping targets and dodging obstacles. The design parameters include weights for exponential control laws for influencing the thrust vector of the drones based on proximity to targets, obstacles and other agents. The overall goal was to decrease R&D time and cost by advancing software prior to implementation. This was one of my favorite projects in college, and I think its a great demonstration of my modeling capabilities. |
|
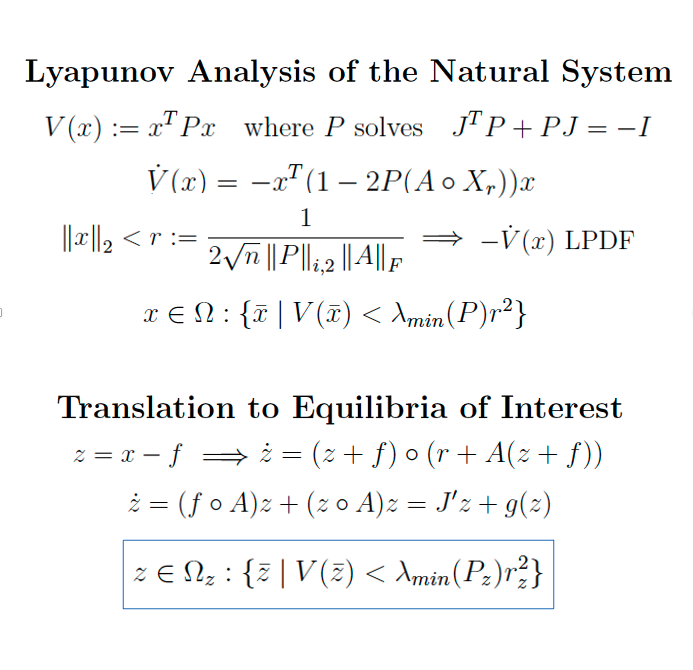
Independent idea, Summer 2020 Derivation / Matlab Implementation Last spring, I took an awesome, graduate course on nonlinear systems and control, and this was an attempt to extend some of the theory to my research. After some wiki-mathing, I decided to use the Hadamard operator to make the algebra more simple. In the end, I proved a bounds for the radius of convergence, which I implemented in matlab. Unfortunately, by this proof, the size of the region is heavily influenced by the Jacobian at a given microbial equilibrium, and in the real examples I have found, this matrix is small. Nonetheless, it sufficed to guarantee convergence of simulated trajectories my colleagues were troubled with. |
|
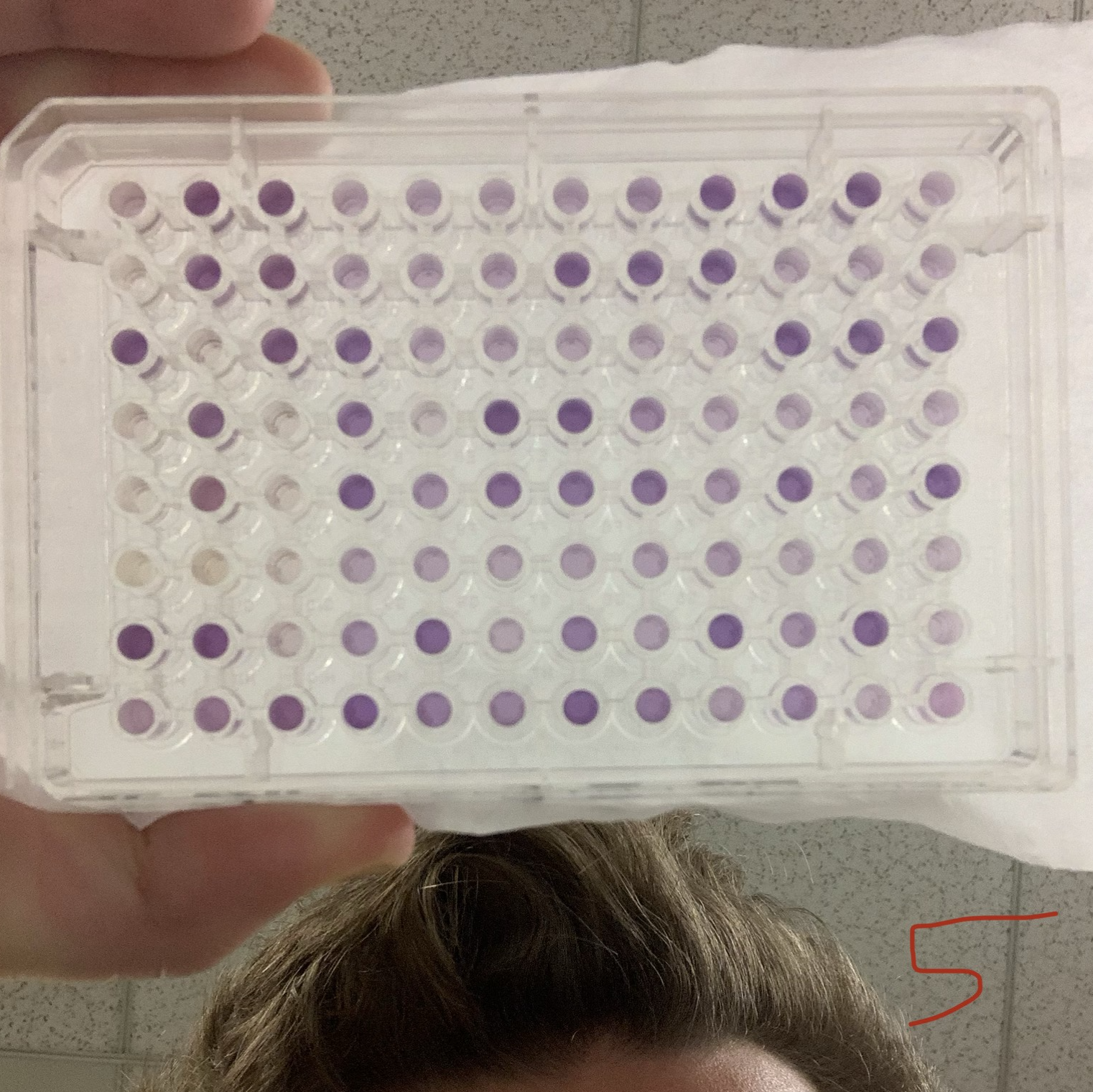
Independent idea, Fall 2019 Matlab script / Validation Figure I took a microbiology class once in which we had to score 8, 96-well biolog plates by hand. I took this as an opportunity to learn matlab's image processing toolbox. The written program takes in phone images, fixes the plane skew of the plate based on user defined corners, isolates the well, attributes the proper biolog label, and takes its average color in cieLAB. Finally, it clusters the found color values and assigns them a score, and returns a list and validation figure. |

|
3rd place in the UC Big Ideas competition, Spring 2019 Proposal I co-lead a team with Ryan Keneally in the 2019 Big Ideas competition to win 3rd place with our proposal to use directed evolution of the AlkB enzyme from a pseudomonad isolate reported to degrade polyethylene. To investigate the proof of concept, Ryan and I also wrote and were awarded two additional grants from the CNR-SPUR and Reagent's funds. Along with writing, I oversaw the genetics. I fused the partially published sequence with a related AlkB and synthesized the sequence with twist. I amplified the fragment to create flanking golden-gate sites and assembled the vector with a pUC vector backbone. Finally, I transformed it into E. coli, P. putida and two P. fluorescens strains for our proof of concept validation. |
|
Thank you Jon Barron for the website template! Feel free to steal this website's source code.
|